Lists¶
Creating Lists¶
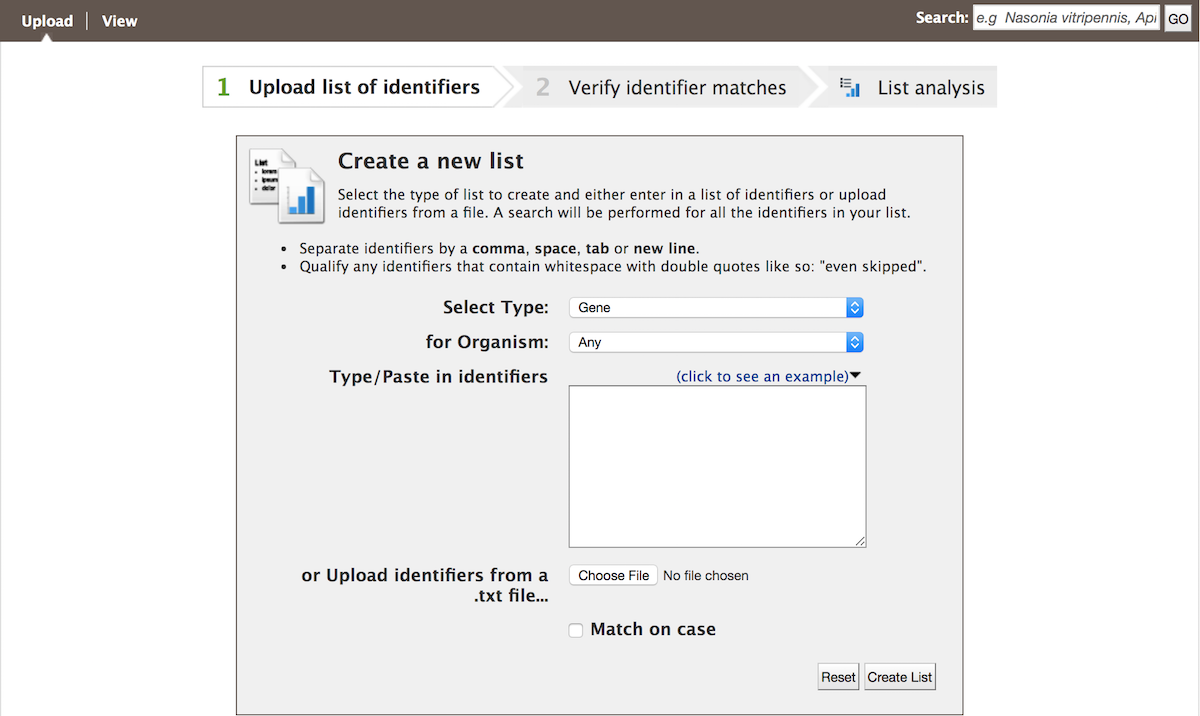
Users may create and save lists of features, such as gene IDs, transcript IDs, gene symbols, etc. The list tool searches the database for the list items and attempts to convert each identifier to the selected type. Click on the Lists tab from the menu to access the full list upload form. A short version of the form is also in the Quick List box on the home page.
As an example, enter the following identifiers (comma-separated):
GB41586, Sec61Beta, TRAM, Mocs1, mal
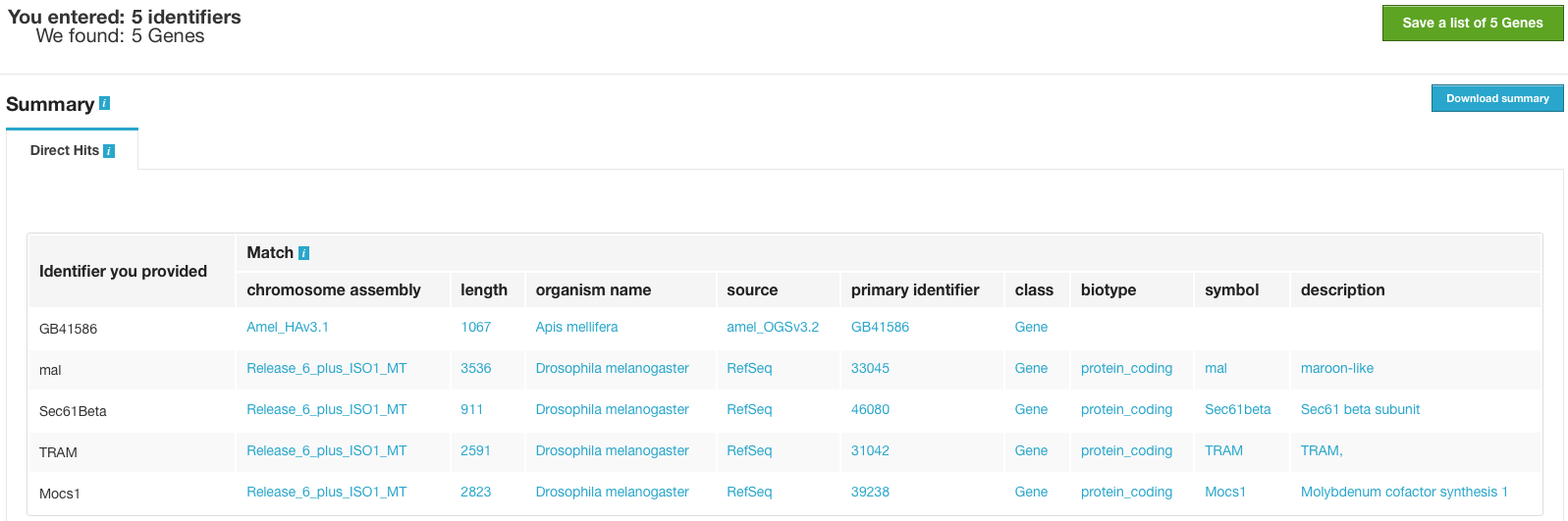
Leave the Select Type as “Gene” and Organism drop-down as “Any”. Then click Create List. A Summary table is displayed with the results of searching for each of the five identifiers in the list.
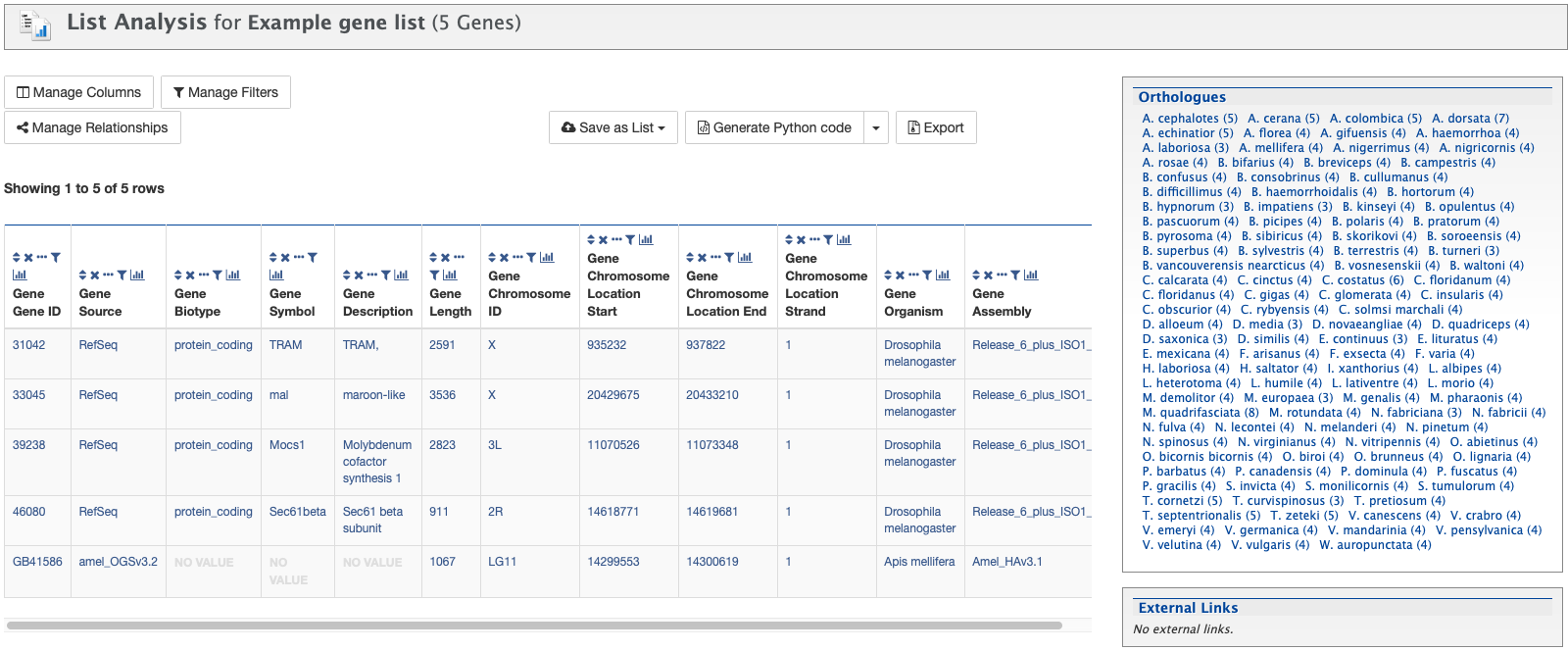
Next, click Save a list of 5 Genes. A List Analysis page is presented that contains widgets allowing users to perform analyses on the genes in the list.
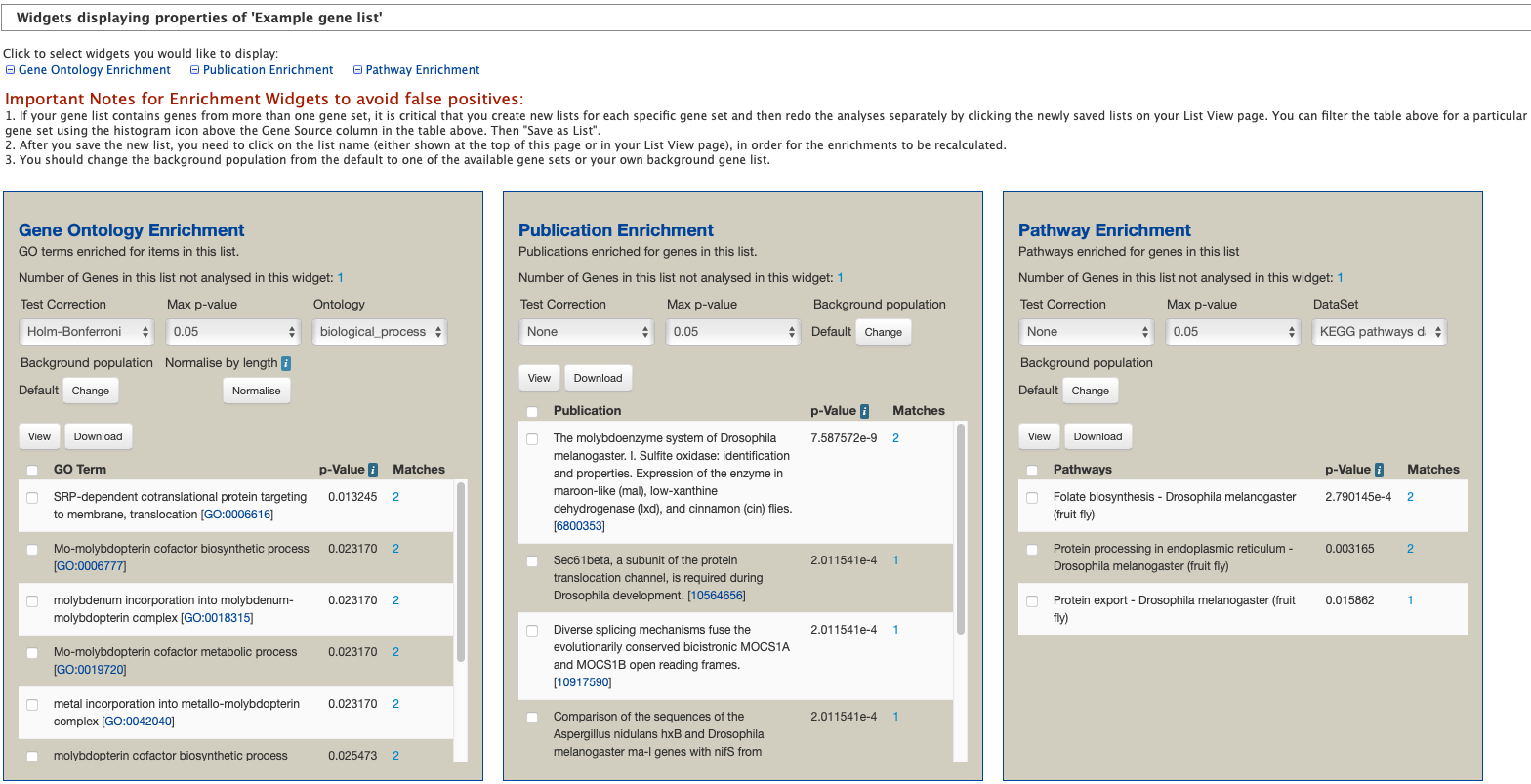
The available widgets are Gene Ontology Enrichment, Publication Enrichment, and Pathway Enrichment. Read the Important Notes for Enrichment Widgets for special instructions to avoid false positives.
Saving Lists¶
Saved lists appear under the View tab on the Lists page. For users who are not logged in, lists are saved temporarily; users must log in to save lists permanently. Saved lists may also be accessed from the MyMine menu item.
Predefined lists of all genes from different species are also available on the Lists page for all users.