Report Pages¶
Every object (e.g., Gene, Protein, Exon) in HymenopteraMine has a report page. The layout of the report page depends on the data available for the object. Report pages may be accessed by clicking on an object name in the results table after running a query.
As an example, on the home page of HymenopteraMine, click on the Protein tab in the Popular Templates section. (Refer to the Templates section for more details on using templates to search the database.) Click on the Gene Symbol –> Proteins template. In the text field, enter LCCH3, and select A. mellifera as the Organism. Then click Show Results.
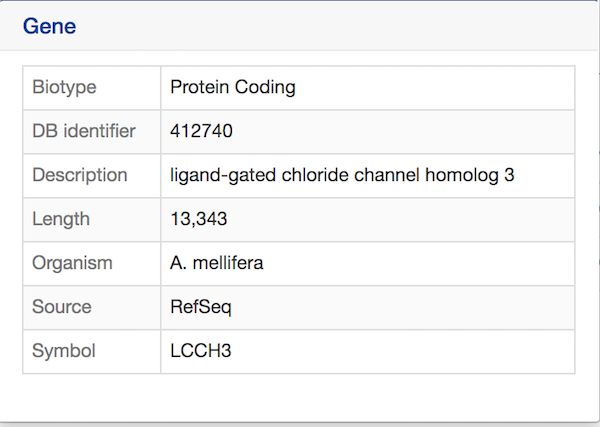
Notice that each item in the results table is a hyperlink. Hover over an item to bring up a quick summary window for that item. For example, hover over LCCH3 to view a summary of the gene with this symbol. The summary contains the gene’s biotype, database identifier, description, length, organism, symbol, and source. Similarly, hover over Q0GQR5_APIME to view a summary of the protein with this DB Identifier.
Clicking on an item in the table rather than just hovering over it will bring up its report page. For example, click on LCCH3 in the Gene Symbol column to view its report.
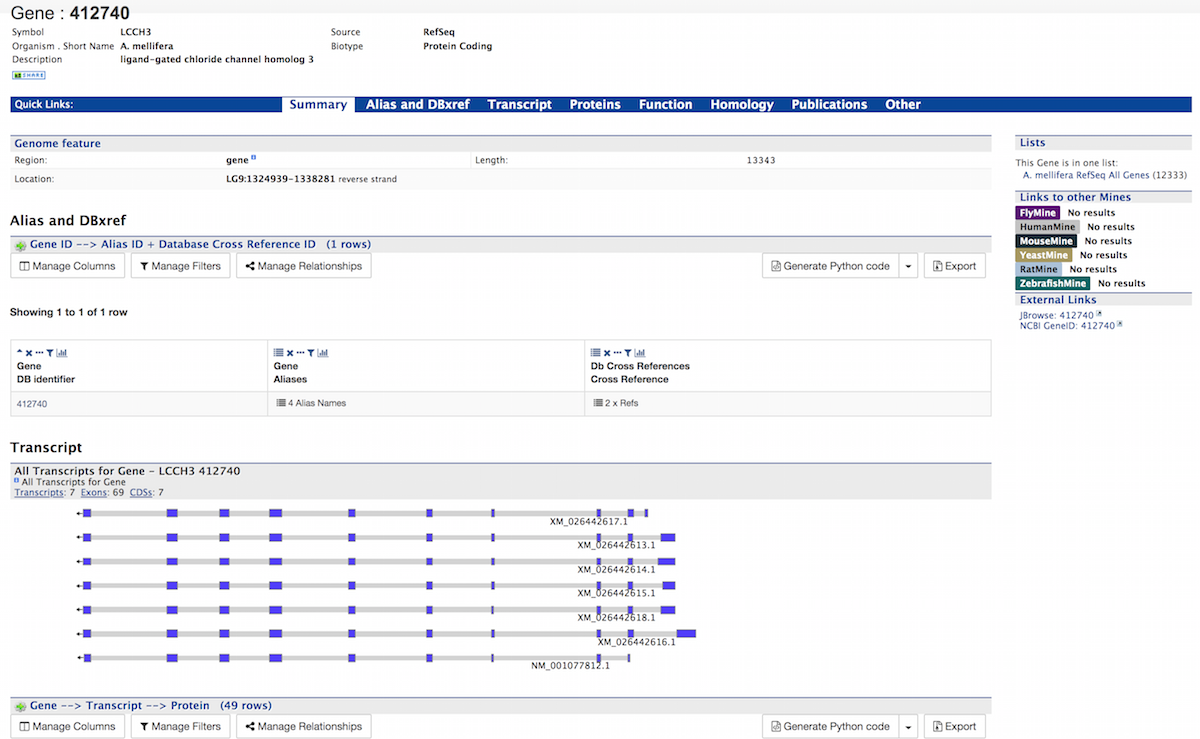
The report page provides a complete description for this gene. The header displays the database identifier, followed by the information from the summary window for the gene (organism, symbol, source, etc.) Biotype indicates the type of gene; in this case the type is protein coding.
The contents of the report page are divided into categories based on the type of information provided.
Summary¶
A Summary section near the top of the report provides information on the gene such as its length, chromosome location, and strand information.

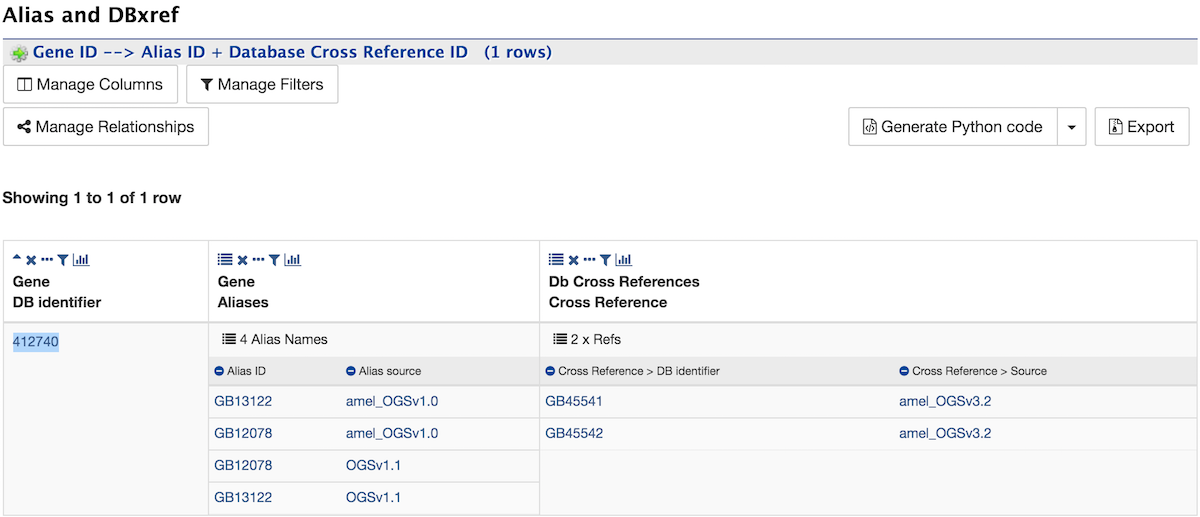
Alias and DBxref¶
The Alias and DBxref section displays a table of aliases and database cross references for the gene. In this example, the gene with DB Identifier 412740 has four aliases and two cross references. Click on the text 4 Alias Names and 2 x Refs to expand the table with additional rows containing the ID and Source for each alias and DB Identifier and Source for each cross reference.
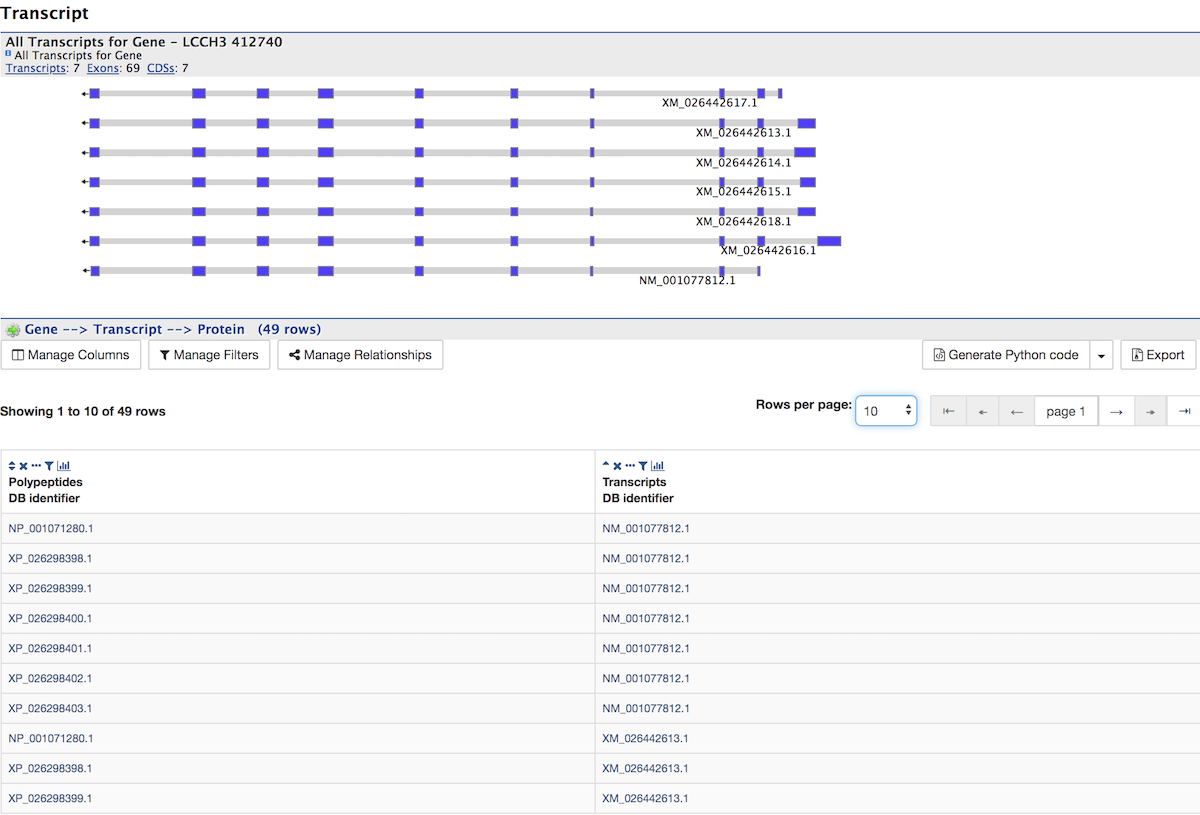
Transcript¶
The Transcript section contains information about the gene model, such as transcripts, exons, etc. It includes a diagram visually representing each transcript with its features highlighted (if applicable). In the case of protein coding genes, a table with protein information is also provided.
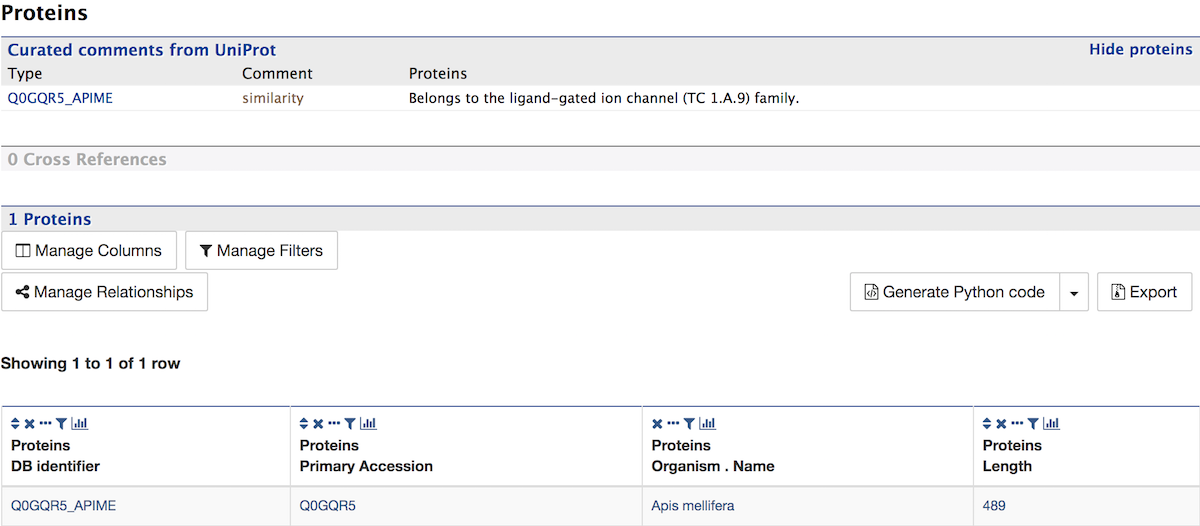
Proteins¶
The Proteins section provides information about the protein product of gene. The comments section gives a brief description about the protein along with the UniProt accession.
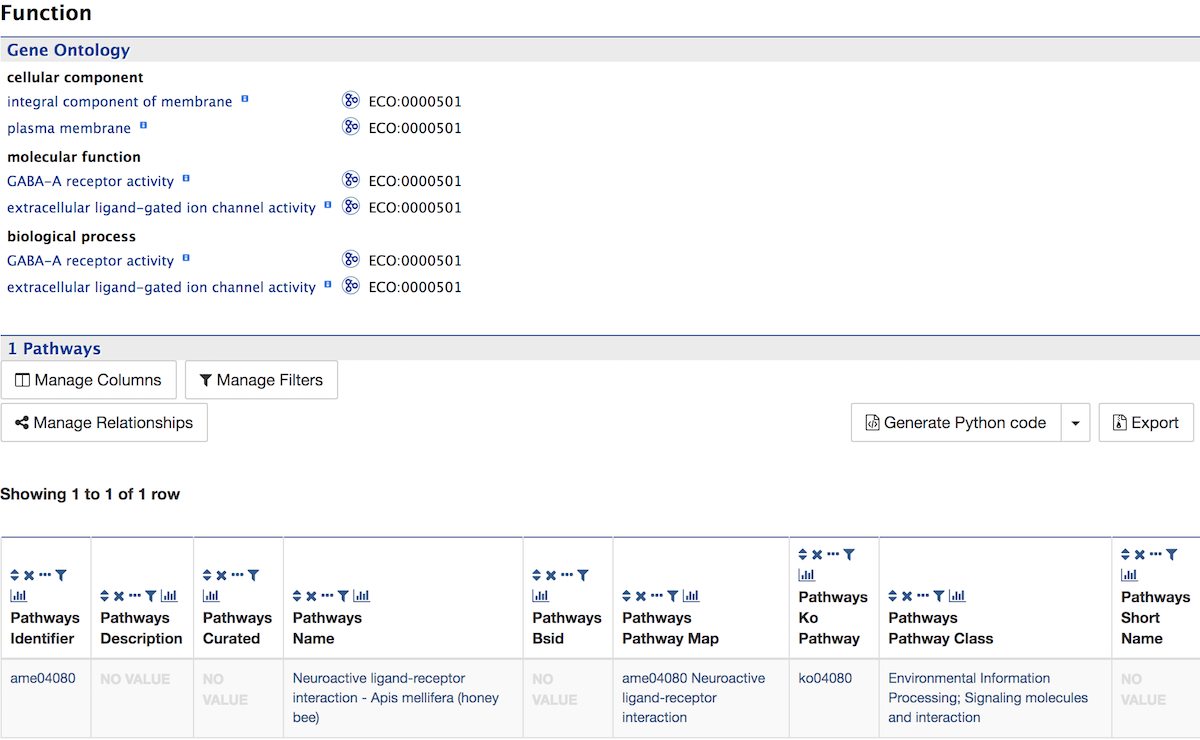
Function¶
The Function section displays Gene Ontology annotations for a gene. Annotations are divided into three categories:
- Cellular Component
- Molecular function
- Biological process
The GO terms are displayed along with the evidence code indicating how the annotations were derived. If applicable, a table of information on Pathways is also shown.
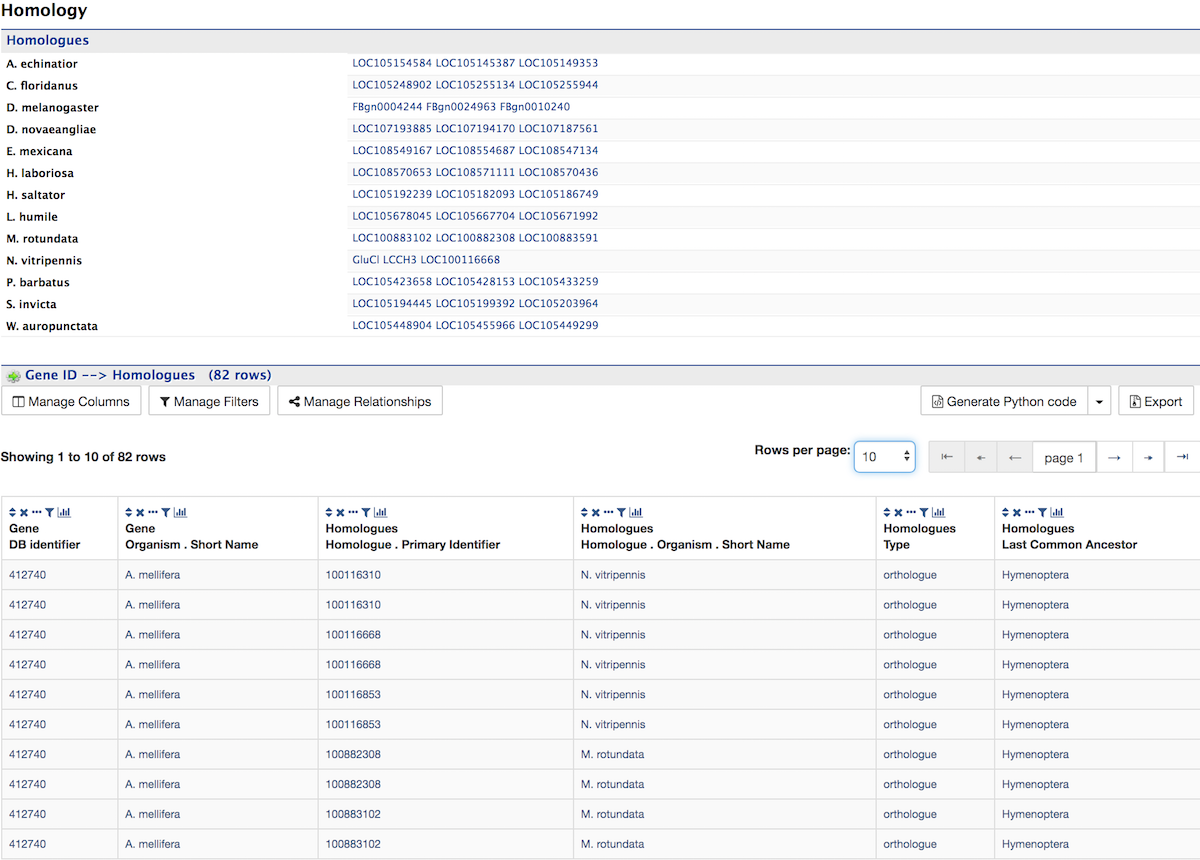
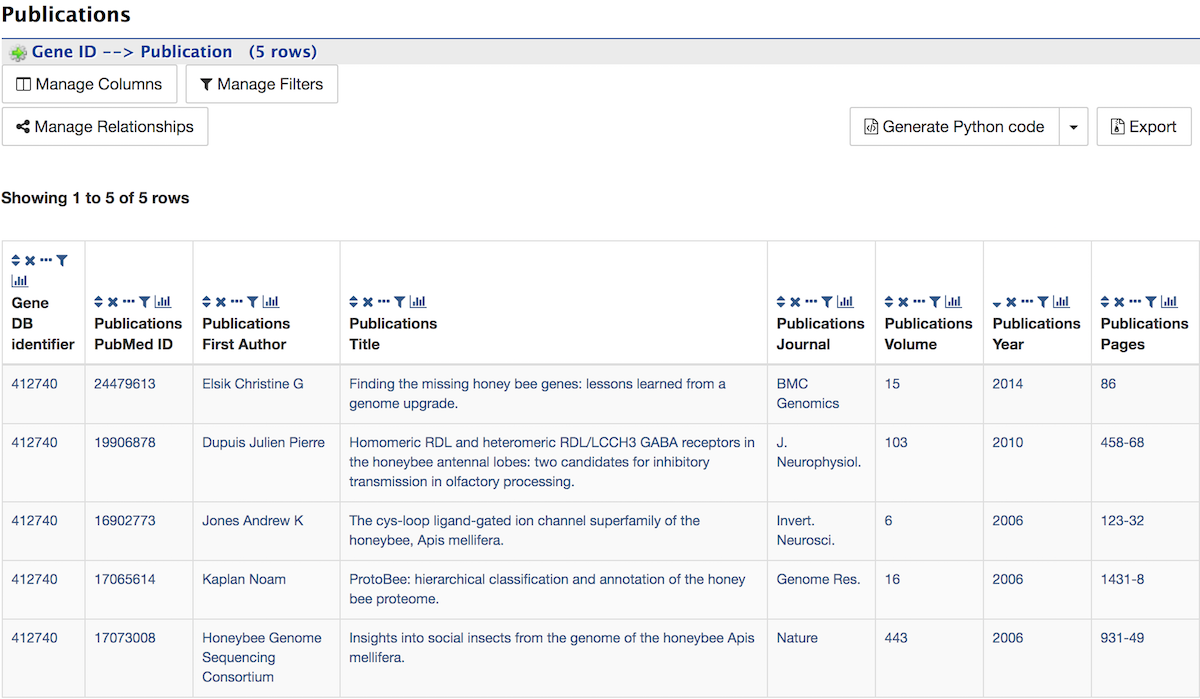
Other¶
This last section provides miscellaneous information that doesn’t fit into any of the above categories, e.g., data sets including a gene, protein domain regions for a protein, etc.